
Mine İlayda Şengör Aygün¹, Özben Yalçın¹, Gülfize Coşkun², Veysel Olgun², FikretDirilenoğlu³, Merve Doğan Ayan¹, Ezgi Toptaş¹, Sıla Özlem Aktaş Şenkaya¹, Işınbike Girente⁴, Cem Çomunoğlu¹.
¹T.C. İstanbul Prof. Dr. Cemil Taşcıoğlu City Hospital, Department of Pathology, Istanbul, Türkiye
²Technomind Dijital Sistemler, BÜDOTEK Teknopark, Istanbul, Türkiye
³Near East University Faculty of Medicine, Department of Pathology, Nicosia, Cyprus
⁴Virasoft, New York, ABD
Introduction:
Compressing original pathology images is essential to reduce storage space requirements. This study examines the impact of compressing PD-L1 (Programmed Death-Ligand 1) stained pathology images using OptiSize software on diagnostic accuracy. The aim of this study is to determine whether there are diagnostic differences between compressed and original images and to evaluate whether necrotic areas, inflammatory cells, and tumor regions can be accurately identified in both image types.
Materials and Methods:
In this study, 52 PD-L1-stained WSI images of cervix, colon, and lung carcinomas with staining percentages ranging from 5 to 95 were used. The original images were compressed using OptiSize Technomind software (Technomind Digital Systems) and then randomly shuffled to be prepared for analysis. Pathologists examined the images in a blinded manner, without knowing which images were original or compressed, and recorded their findings in an Excel spreadsheet. The pathologists assessed their ability to accurately identify percentage scores, necrotic areas, inflammatory cells, and tumor regions in both sets of images. To mathematically evaluate the quality of the images, pixel-based PSNR and structural-based SSIM metrics were used. While SSIM measures structural similarities, PSNR assesses noise levels relative to the maximum pixel value of the images. When used together, these methods provide a comprehensive analysis of the quality and similarity of the images. Additionally, the images were analyzed using the QuPath image analysis software.
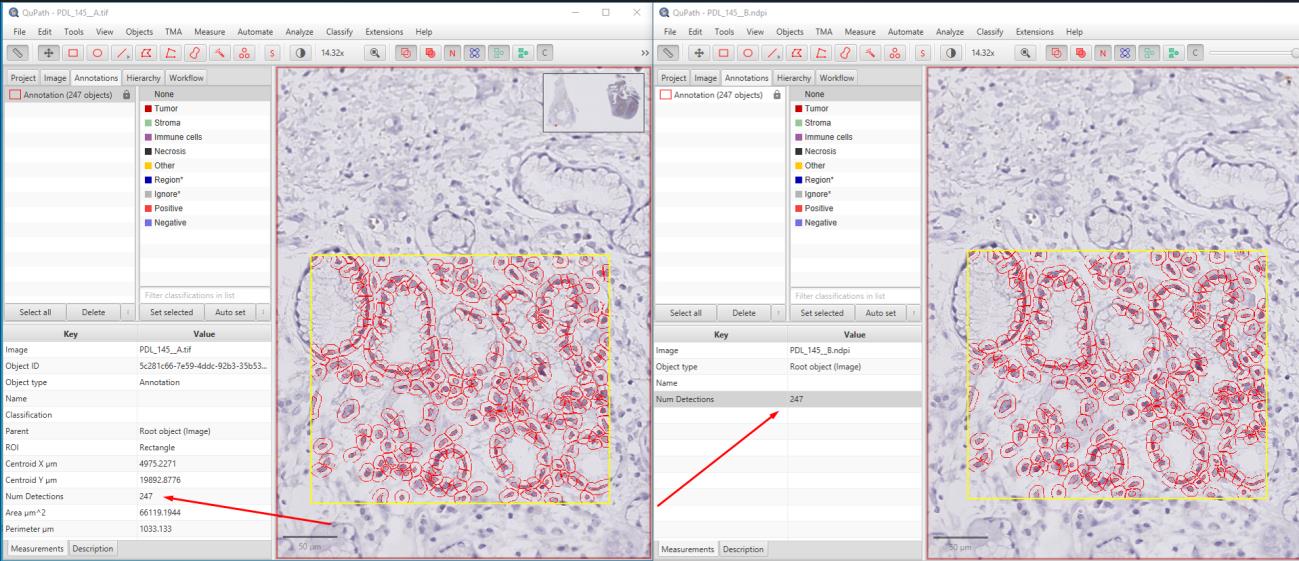
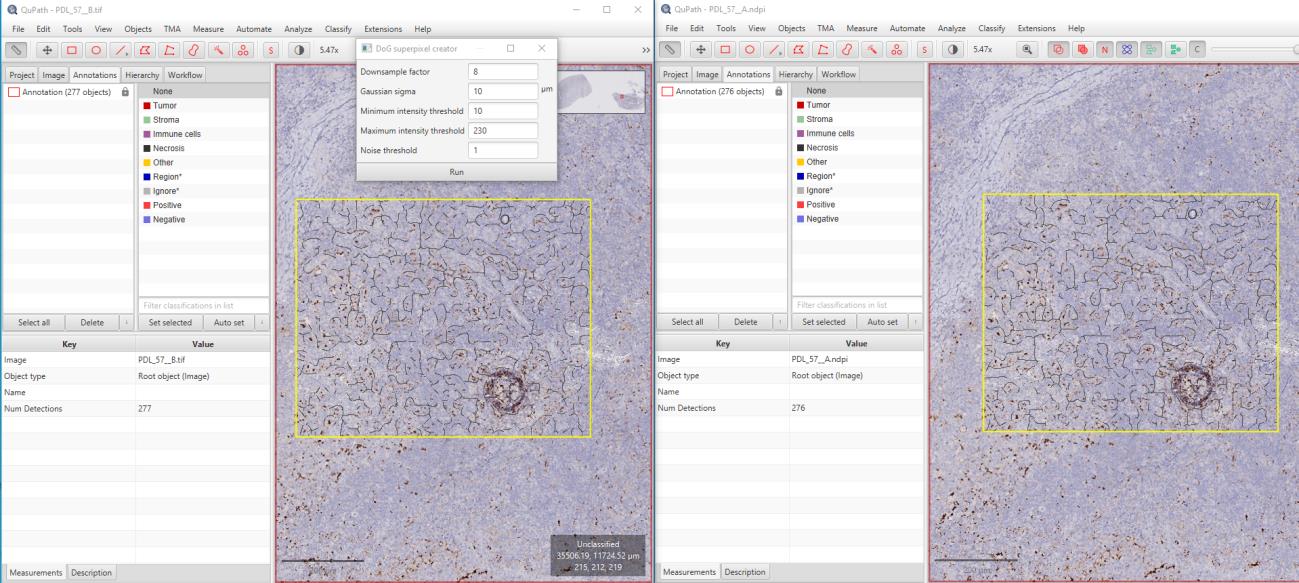
Results:
The pathologists' evaluations revealed no elements affecting the diagnosis between the original and compressed images. It was observed that necrotic areas, inflammatory cells, and tumors could be accurately identified in both image sets, and the percentage scores were given similarly for both image types. The original images, which totaled 127 GB, were compressed to 61.6 GB using OptiSize. The similarity index between the images was calculated using the SSIM and PSNR methods, yielding an average value of 0.984 (min: 0.981, max: 0.990) for the 52 images. The PSNR method produced an average value of 40.79 dB (min: 40.021, max: 41.789). These results indicate that the images maintained high quality with minimal degradation.
Conclusion:
It was observed that compression using the OptiSize WSI compression algorithm did not affect the staining result in the immunohistochemical PD-L1 evaluation, and there was no visible difference affecting the diagnosis in both image sets. Additionally, the evaluations conducted using QuPath yielded highly correlated results. Thus, with the ability to store high-volume WSI images with up to 55% savings, it is possible to reduce costs and increase the data storage capacity of laboratories. These results demonstrate that the OptiSize algorithm is a safe compression method for pathological evaluations. Keywords: Whole Slide Imaging (WSI), Image Compression, PD-L1, Immunohistochemistry, Pathology