
Comparison of Original and Compressed Pathology Images in Helicobacter Pylori Diagnosis
Özben Yalçın¹, Gülfize Coşkun², Veysel Olgun², Emre Şenkaya¹, Fatma Korkmaz Aydın¹, Emel Cingi Doğani¹, Nazlı Ecem Kore¹, Seden Atike¹, Arsoy Şahin¹, Refik Selim Atamanalp¹, Buse Akı¹, Cem Çomunoğlu¹
¹T.C. İstanbul Prof. Dr. Cemil Taşcıoğlu City Hospital, Department of Pathology, Istanbul, Türkiye
²Technomind Dijital Sistemler, BÜDOTEK Teknopark, Istanbul, Türkiye
Introduction:
The compression of digital pathology images is becoming increasingly important to facilitate data storage and management processes. This study examined the effects of compression using OptiSize software on the diagnostic accuracy of Helicobacter-stained pathology images. We investigated how compression affected diagnostic sensitivity and determined whether there are any diagnostic differences between compressed and original images.
Materials and Methods:
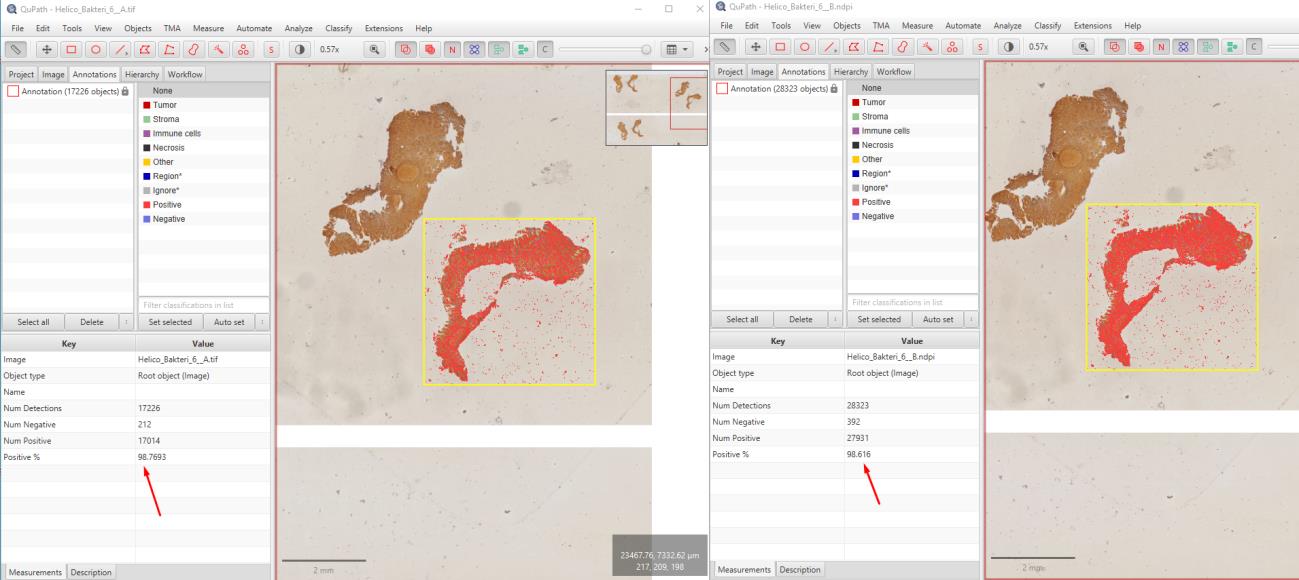
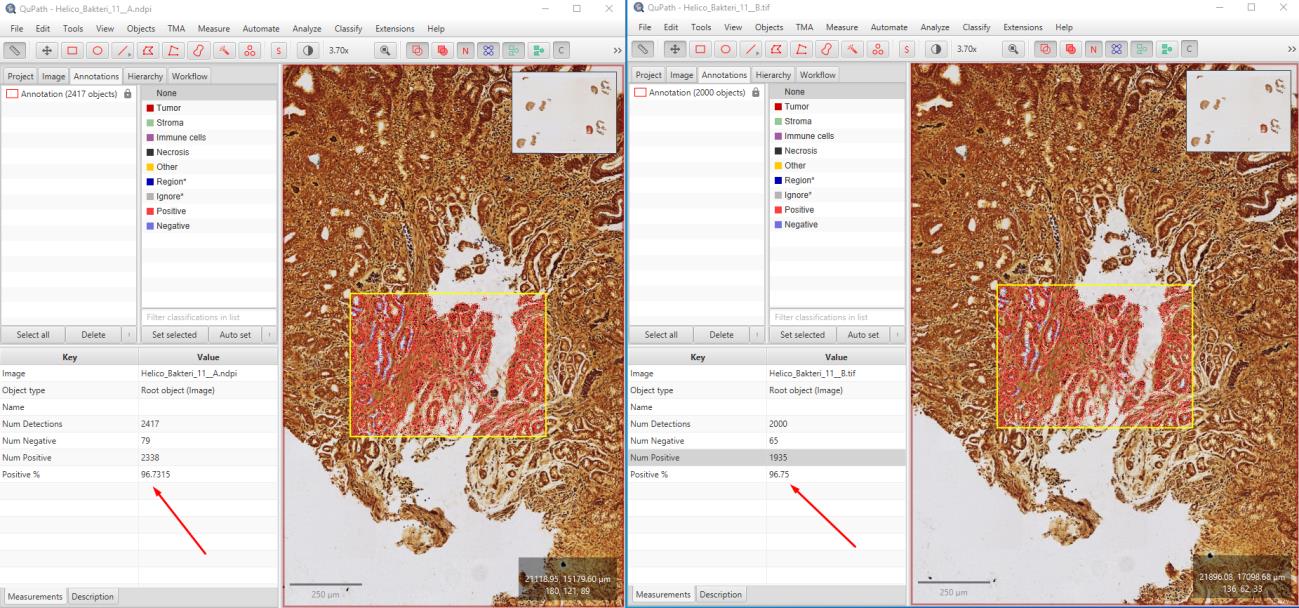
In this study, 115 immunohistochemically Helicobacter-stained WSI images of gastric biopsies were compressed using OptiSize. The images were graded as negative, +1, +2, and +3. Pathologists assessed the Helicobacter grading on a total of 170 (85 original and 85 compressed) images and examined 60 images (30 original and 30 compressed) for synchronized differences. These assessments were recorded in an Excel spreadsheet. The study was conducted in a blinded manner by randomly mixing the original and compressed images labeled as A and B. To evaluate the image quality, calculations were performed using the SSIM and PSNR methods, and analyses were conducted on the images using the open-source QuPath image analysis software.
Results:
The evaluation revealed that compressing Helicobacter images with OptiSize had no adverse effect on diagnostic accuracy. The Helicobacter grades (negative, +1, +2, +3) were consistently determined in both the original and compressed images. In this blinded study, the Helicobacter grading was the same for both the original and compressed images across a total of 170 images.The total size of the original images, which was 115 GB, was reduced to 59 GB using the OptiSize software. Mathematical calculations for 118 images resulted in an average SSIM value of 0.983 (min: 0.980, max: 0.990). The PSNR method yielded an average value of 39.92 dB (min: 38.17, max: 42.167). Analyses conducted with QuPath showed correlated results in the count of positive cells.
Conclusion:
The study demonstrates that the OptiSize WSI compression algorithm does not affect diagnostic accuracy in Helicobacter immunohistochemical analyses. Pathologists reported no diagnostic differences between the original and compressed images. These findings indicate that compressing high-volume WSI images is an effective way to enhance data storage capacity and reduce costs in laboratories. The study suggests that the OptiSize algorithm can be reliably used as a compression method in pathological evaluations, contributing to the broader adoption of digital pathology in clinical and research settings. Future studies will include more comprehensive analyses using advanced artificial intelligence tools.
Keywords: WSI, Image Compression, Helicobacter, Immunohistochemistry, Digital Pathology